|
Immunohistochemistry
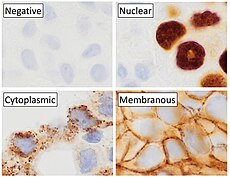   Immunohistochemistry is a form of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells and tissue, by exploiting the principle of antibodies binding specifically to antigens in biological tissues. Albert Hewett Coons, Ernest Berliner, Norman Jones and Hugh J Creech was the first to develop immunofluorescence in 1941. This led to the later development of immunohistochemistry.[2][3] Immunohistochemical staining is widely used in the diagnosis of abnormal cells such as those found in cancerous tumors. In some cancer cells certain tumor antigens are expressed which make it possible to detect. Immunohistochemistry is also widely used in basic research, to understand the distribution and localization of biomarkers and differentially expressed proteins in different parts of a biological tissue.[4] Sample preparationImmunohistochemistry can be performed on tissue that has been fixed and embedded in paraffin, but also cryopreservated (frozen) tissue. Based on the way the tissue is preserved, there are different steps to prepare the tissue for immunohistochemistry, but the general method includes proper fixation, antigen retrieval incubation with primary antibody, then incubation with secondary antibody.[5][6] Tissue preparation and fixationFixation of the tissue is important to preserve the tissue and maintaining cellular morphology. The fixation formula, ratio of fixative to tissue and time in the fixative, will affect the result. The fixation solution (fixative) is often 10% neutral buffer formalin. Normal fixation time is 24 hours in room temperature. The ratio of fixative to tissue ranges from 1:1 to 1:20. After the tissue is fixed it can be embedded in paraffin wax.[5][6] For frozen sections, fixation is usually performed after sectioning if not new antibodies are going to be tested. Then acetone or formalin can be used. [6] SectioningSectioning of the tissue sample is done using a microtome. For paraffin embedded tissue 4 μm is normal thickness, and for frozen sections 4 – 6 μm.[6] The thickness of the sliced sections matters, and is an important factor in immunohistochemistry. If you compare a section of brain tissue measuring 4 μm with a section measuring 7 μm, some of what you see in the 7 μm thick section might be lacking in the 4 μm section. This shows the importance of detailed methods related to this methodology.[7] The paraffin embedded tissues should be deparaffinized to remove all the paraffin on and around the tissue sample in xylene or a good substitute, followed by alcohol.[8] Antigen retrievalAntigen retrieval is required to make the epitopes accessible for immunohistochemical staining for most formalin fixed tissue section. The epitopes are the binding sites for antibodies used to visualize the targeted antigen which may be masked due to the fixation. Fixation of the tissue may cause formation of methylene bridges or crosslinking of amino groups, so that the epitopes no longer are available. Antigen retrieval can restore the masked antigenicity, possibly by breaking down the crosslinks caused by fixation. [9] The most common way to perform antigen retrieval is by using high-temperature heating while soaking the slides in a buffer solution. [10] This can be done in different ways, for example by using microwave oven, autoclaves, heating plates or water baths. For frozen sections, antigen retrieval is generally not necessary, but for frozen section that has been fixed in acetone or formalin, can antigen retrieval improve the immunohistochemistry signal.[6] BlockingNon-specific binding of antibodies can cause background staining. Although antibodies bind to specific epitopes, they may also partially or weakly bind to sites on nonspecific proteins that are similar to the binding site on the target protein. By incubating the tissue with normal serum isolated from the species which the secondary antibody was produced, the background staining can be reduced. It is also possible to use commercially available universal blocking buffers. Other common blocking buffers include normal serum, non-fat dry milk, BSA, or gelatin.[5][6] Endogenous enzyme activity may also cause background staining but can be reduced if the tissue is treated with hydrogen peroxide.[5] Sample labelingAfter preparing the sample, the target can be visualized by using antibodies labeled with fluorescent compounds, metals or enzymes. There are direct and indirect methods for labeling the sample.[6][11] Antibody typesThe antibodies used for detection can be polyclonal or monoclonal. Polyclonal antibodies are made by using animals like guinea pig, rabbit, mouse, rat, or goat. The animal is injected with the antigen of interest and trigger an immune response. The antibodies can be isolated from the animal's whole serum. Polyclonal antibody production will result in a mixture of different antibodies and will recognize multiple epitopes. Monoclonal antibodies are made by injecting the animal with the antigen of interest and then isolating an antibody-producing B cell, typically from the spleen. The antibody producing cell is then fused with a cancer cell line. This causes the antibodies to show specificity for a single epitope.[12] For immunohistochemical detection strategies, antibodies are classified as primary or secondary reagents. Primary antibodies are raised against an antigen of interest and are typically unconjugated (unlabeled). Secondary antibodies are raised against immunoglobulins of the primary antibody species. The secondary antibody is usually conjugated to a linker molecule, such as biotin, that then recruits reporter molecules, or the secondary antibody itself is directly bound to the reporter molecule.[11] Detection methodsThe direct method is a one-step staining method and involves a labeled antibody reacting directly with the antigen in tissue sections. While this technique utilizes only one antibody and therefore is simple and rapid, the sensitivity is lower due to little signal amplification, in contrast to indirect approaches.[11] The indirect method involves an unlabeled primary antibody that binds to the target antigen in the tissue. Then a secondary antibody, which binds with the primary antibody is added as a second layer. As mentioned, the secondary antibody must be raised against the antibody IgG of the animal species in which the primary antibody has been raised. This method is more sensitive than direct detection strategies because of signal amplification due to the binding of several secondary antibodies to each primary antibody.[11] The indirect method, aside from its greater sensitivity, also has the advantage that only a relatively small number of standard conjugated (labeled) secondary antibodies needs to be generated. For example, a labeled secondary antibody raised against rabbit IgG, is useful with any primary antibody raised in rabbit. This is particularly useful when a researcher is labeling more than one primary antibody, whether due to polyclonal selection producing an array of primary antibodies for a singular antigen or when there is interest in multiple antigens. With the direct method, it would be necessary to label each primary antibody for every antigen of interest.[11]  Reporter moleculesReporter molecules vary based on the nature of the detection method, the most common being chromogenic and fluorescence detection. In chromogenic immunohistochemistry an antibody is conjugated to an enzyme, such as alkaline phosphate and horseradish peroxidase, that can catalyze a color-producing reaction in the presence of a chromogenic substrate like diaminobenzidine.[5] The colored product can be analyzed with an ordinary light microscope.[13] In immunofluorescence the antibody is tagged to a fluorophore, such as fluorescein isothiocyanate, tetramethylrhodamine isothiocyanate, aminomethyl Coumarin acetate or Cyanine5. Synthetic fluorochromes from Alexa Fluors is also commonly used.[13][14] The fluorochromes can be visualized by a fluorescence or confocal microscope.[13] For chromogenic and fluorescent detection methods, densitometric analysis of the signal can provide semi- and fully quantitative data, respectively, to correlate the level of reporter signal to the level of protein expression or localization.[6] Counterstains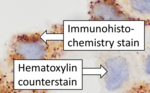 After immunohistochemical staining of the target antigen, another stain is often applied. The counterstain provide contrast that helps the primary stain stand out and makes it easier to examine the tissue morphology. It also helps with orientation and visualization of the tissue section. Hematoxylin is commonly used.[6][15] TroubleshootingIn immunohistochemical techniques, there are several steps prior to the final staining of the tissue that can cause a variety of problems. It can be strong background staining, weak target antigen staining and presence of artifacts. It is important that antibody quality and the immunohistochemistry techniques are optimized.[16] Endogenous biotin, reporter enzymes or primary/secondary antibody cross-reactivity are common causes of strong background staining.[11][13] Weak or absent staining may be caused by inaccurate fixation of the tissue or to low antigen levels. These aspects of immunohistochemistry tissue prep and antibody staining must be systematically addressed to identify and overcome staining issues.[5][6]  Methods to eliminate background staining include dilution of the primary or secondary antibodies, changing the time or temperature of incubation, and using a different detection system or different primary antibody. Quality control should as a minimum include a tissue known to express the antigen as a positive control and negative controls of tissue known not to express the antigen, as well as the test tissue probed in the same way with omission of the primary antibody (or better, absorption of the primary antibody).[5] [18] Diagnostic immunohistochemistry markers Immunohistochemistry is an excellent detection technique and has the tremendous advantage of being able to show exactly where a given protein is located within the tissue examined. It is also an effective way to examine the tissues. This has made it a widely used technique in neuroscience, enabling researchers to examine protein expression within specific brain structures. Its major disadvantage is that, unlike immunoblotting techniques where staining is checked against a molecular weight ladder, it is impossible to show in immunohistochemistry that the staining corresponds with the protein of interest. For this reason, primary antibodies must be well-validated in a Western Blot or similar procedure. The technique is even more widely used in diagnostic surgical pathology for immunophenotyping tumors (e.g. immunostaining for e-cadherin to differentiate between ductal carcinoma in situ (stains positive) and lobular carcinoma in situ (does not stain positive)[19]). More recently, immunohistochemical techniques have been useful in differential diagnoses of multiple forms of salivary gland, head, and neck carcinomas.[20] The diversity of immunohistochemistry markers used in diagnostic surgical pathology is substantial. Many clinical laboratories in tertiary hospitals will have menus of over 200 antibodies used as diagnostic, prognostic and predictive biomarkers. Examples of some commonly used markers include:
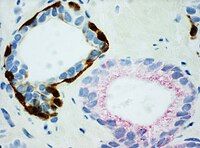
Directing therapyA variety of molecular pathways are altered in cancer and some of the alterations can be targeted in cancer therapy. Immunohistochemistry can be used to assess which tumors are likely to respond to therapy, by detecting the presence or elevated levels of the molecular target.[citation needed] Chemical inhibitorsTumor biology allows for a number of potential intracellular targets. Many tumors are hormone dependent. The presence of hormone receptors can be used to determine if a tumor is potentially responsive to antihormonal therapy. One of the first therapies was the antiestrogen, tamoxifen, used to treat breast cancer. Such hormone receptors can be detected by immunohistochemistry.[23] Imatinib, an intracellular tyrosine kinase inhibitor, was developed to treat chronic myelogenous leukemia, a disease characterized by the formation of a specific abnormal tyrosine kinase. Imitanib has proven effective in tumors that express other tyrosine kinases, most notably KIT. Most gastrointestinal stromal tumors express KIT, which can be detected by immunohistochemistry.[24] Monoclonal antibodiesMany proteins shown to be highly upregulated in pathological states by immunohistochemistry are potential targets for therapies utilising monoclonal antibodies. Monoclonal antibodies, due to their size, are utilized against cell surface targets. Among the overexpressed targets are members of the EGFR family, transmembrane proteins with an extracellular receptor domain regulating an intracellular tyrosine kinase.[25] Of these, HER2/neu (also known as Erb-B2) was the first to be developed. The molecule is highly expressed in a variety of cancer cell types, most notably breast cancer. As such, antibodies against HER2/neu have been FDA approved for clinical treatment of cancer under the drug name Herceptin. There are commercially available immunohistochemical tests, Dako HercepTest, Leica Biosystems Oracle[26] and Ventana Pathway.[27] Similarly, epidermal growth factor receptor (HER-1) is overexpressed in a variety of cancers including head and neck and colon. Immunohistochemistry is used to determine patients who may benefit from therapeutic antibodies such as Erbitux (cetuximab).[28] Commercial systems to detect epidermal growth factor receptor by immunohistochemistry include the Dako pharmDx. Mapping protein expressionImmunohistochemistry can also be used for a more general protein profiling, provided the availability of antibodies validated for immunohistochemistry. The Human Protein Atlas displays a map of protein expression in normal human organs and tissues. The combination of immunohistochemistry and tissue microarrays provides protein expression patterns in a large number of different tissue types. Immunohistochemistry is also used for protein profiling in the most common forms of human cancer.[29][30] See also
References
Further reading
External linksWikimedia Commons has media related to Immunohistochemistry.
|